The cannabinoid receptor type 1
The cannabinoid receptor type 1 (CB1R) is a G-protein coupled receptor (GPCR), abundant in the central nervous system, that is activated by the key psychoactive component in Cannabis species, delta-9-tetrahydrocannabinol (THC).1 Beyond its well-described role in psychopharmacology, CB1R is also strongly implicated in regulating food intake, and inappropriate CB1R signalling contributes to the development of obesity and other metabolic conditions, especially type 2 diabetes.2,3 Indeed, an inverse agonist of CB1R (a compound that produces the opposite signal from a receptor, rather than simply blocking it), rimonabant, was briefly marketed as an anti-obesity and anti-diabetic drug in the late 2000s. However, rimonabant was withdrawn worldwide scant years later amid serious concerns over its safety prompted by unexpected psychiatric side effects.4
Allosteric modulators
Allosteric modulators are compounds that bind to receptors, such as CB1R, at a different place than the compound or hormone our bodies produce to interact with the receptor (known as the orthosteric site).5,6 In this way, they can enhance (positive allosteric modulation; PAM) or diminish (negative allosteric modulation; NAM) the existing natural signalling of the receptor.7,8 The essential promise of allosteric modulators as drugs is that they can influence the activity of their targets in a more subtle way.
For psychedelic drug research, there are obvious advantages to this approach. Many GPCRs, such as CB1R and the serotonin receptors, produce potentially undesirable psychoactive effects when activated, which complicates the use of agonists (activating drugs that act on the orthosteric site).9 Furthermore, direct inactivation, or inverse agonism, can be dangerous, as seen with rimonabant. Allosteric modulation may therefore offer ways to avoid undesirable “on-target” effects for receptors involved in human responses to psychedelics.
X-ray crystal structure of CB1R with a PAM
In an important paper recently published in Nature Chemical Biology, Yang and colleagues used a combination of cutting-edge techniques to convincingly reveal how allosteric modulators can bind to CB1R and influence its function.5
Firstly, they employed x-ray crystallography to obtain the structure of CB1R (grey) bound simultaneously to a synthetic cannabinoid, CP 55,940, and a positive allosteric modulator, ZCZ011. In this technique, large amounts of purified receptor protein are mixed with one or more drugs of interest, and grown carefully into crystals. When high intensity x-rays are fired through these crystals, they scatter into specific patterns. With the help of computers, scientists can use these patterns to reconstruct the precise 3D structures of the receptor and drugs.
From this experiment, Yang and colleagues found that ZCZ011, the PAM (yellow), inserts into a cleft on one side of the protein, which sits in between three of CB1R’s eight helix domains (pale green) (Figure 1). ZCZ011 pushes parts of these helices inward, towards the receptor’s centre, where agonists bind (Figure 1, right panel). This physically moves amino acids inside the orthosteric site, particularly serine 173 (S173, magenta), closer to the agonist CP 55,940 (blue), improving its binding to CB1R. Indeed, S173 must be important for the activating effect of this synthetic cannabinoid, as CB1R activation by CP 55,940 drops 60-fold when S173 is mutated to a different amino acid.
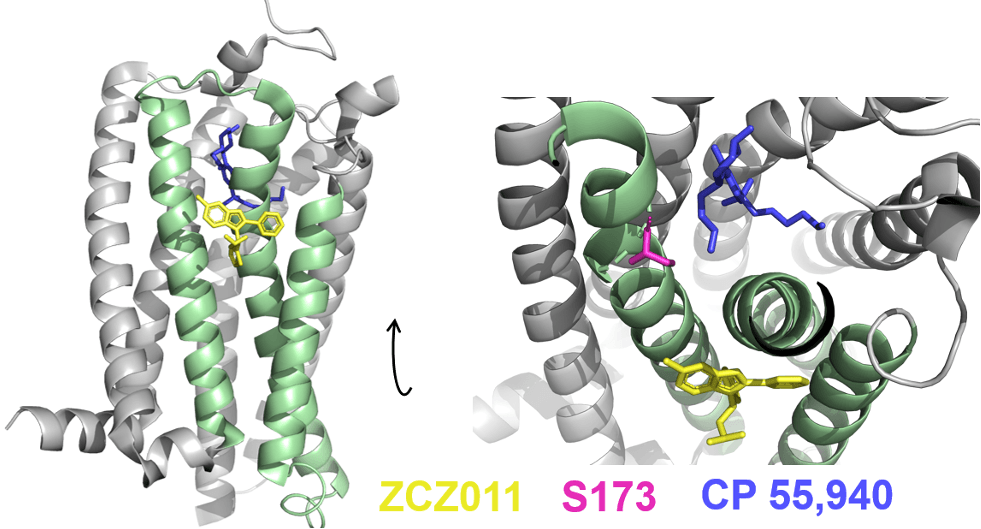
Figure 1: ZCZ011, a PAM, binds in a cleft between helices 2, 3, and 4 (pale green) of CB1R. Inside the orthosteric binding site, an amino acid S173 (magenta) important for activation by the agonist CP 55,940 (pale blue) is pushed closer to the agonist, strengthening its binding. The molecular coordinates for this x-ray crystal structure were obtained from the Protein Databank, PDB: 7FEE.
Cryogenic electron microscopy structure of CB1R with a PAM
It has previously been shown that agonists, such as the synthetic cannabinoid MDMB-Fubinaca, insert deep into the centre of the receptor and disrupt an essential “toggle switch” that physically holds the receptor in an inactive state (Figure 2A).10 Disrupting this switch forces CB1R to undergo a series of shape changes, ultimately leading to a large outward movement of the receptor’s 6th helix. This creates a larger cavity on the side of the receptor inside the cell, allowing G proteins to interact with this side and begin signalling (Figure 2B).

Figure 2: Activation of cannabinoid receptor type 1 by a synthetic cannabinoid. A) CB1R (bright green) is activated when agonists, such as MDMB-Fubinaca (bright orange) bind to its centre. This binding unlocks a critical “toggle switch” (inset, bright green), which normally limits CB1R’s ability to signal. B) A series of shape changes occur after activation, causing CB1R’s 6th helix (yellow) to rotate outward by a large amount when activated (green). The molecular coordinates were obtained from the Protein Databank, PDB: 6N4B (activated CB1R), 6KQI (inactive CB1R).
However, the x-ray crystal structure presented by Yang and colleagues is clearly in the inactive state, based on the inward position of helix 6. This is despite being bound to an agonist and a PAM, which prevents a true understanding of how ZCZ011 changes CB1R structure to achieve positive modulation. To better connect their results to human biology, the authors obtained a second structure of CB1R, fully active and in a complex with its signalling partner G proteins, by cryogenic electron microscopy (cryo-EM) (Figure 3A).
Cryo-EM is a relatively new structural biology technique that takes direct, extremely high magnification images of proteins using specially modified electron microscopes. The purified proteins (and any drugs) are snap-frozen in ice, and the microscopy is done under cooling from liquid nitrogen. Images of millions of proteins are collected, and these can be mathematically averaged on a supercomputer to produce a high-resolution structure with near-atomic details. A particular advantage of cryo-EM over other techniques is that the proteins are captured in an environment very similar to that in our bodies, and it is generally thought that the 3D structures obtained this way are more relevant to human biology. In conjunction with computer simulations, this new cryo-EM structure proved critical for the authors to understand how these allosteric modulators work at CB1R.

Figure 3: The cryo-EM structure of CB1R reveals the mechanism of allosteric modulation. A) Overview of the cannabinoid receptor type 1 (pale green) in a fully activated signalling complex with its partner G proteins (salmon, pale blue, and pale yellow). B) For CB1R to activate, a critical amino acid, F155 (salmon, red) must undergo a large rotation. ZCZ011 (yellow), a PAM, enhances rotation of F155. C) A NAM, ORG27579 (salmon) binds to CB1R further down in the same cleft as ZCZ011 (yellow). It has the opposite effect on F155 (red), making it more difficult to rotate during activation of CB1R by an agonist such as CP 55,940 (blue). The molecular coordinates were obtained from the Protein Databank, PDB: 7WV9 (active CB1R with PAM, cryo-EM), 6KQI (coordinates for ORG27569 aligned onto 7WV9).
Molecular dynamics simulations
However, structures alone are really only snapshots of the movements and actions that receptors go through when drugs interact with them. To enhance their findings, the authors ran extensive molecular dynamics (MD) simulations. The results indicated that a single amino acid of CB1R, phenylalanine 155 (F155) must undergo a substantial rotation in order to enable the activation of CB1R when agonists bind. This is part of a larger series of shape changes that need to happen for CB1R to be activated by agonists. Indeed, in their fully activated cryo-EM structure, this amino acid, F155, had undergone the necessary rotation, whereas in their x-ray crystal structure, it had not (Figure 3B). The cryo-EM structure revealed that ZCZ011 alters the receptor’s shape to make it easier for the amino acid F155 to rotate, enhancing activation by agonists such as CP 55,940 and explaining why ZCZ011 is a positive allosteric modulator.
What about negative allosteric modulation?
Lastly, the authors extended the MD simulations to encompass information obtained from a previous x-ray crystal structure with a negative allosteric modulator, ORG27569.11 Curiously, this NAM binds in the same cleft as the PAM, ZCZ011 (Figure 3C). In this position, ORG27569 (salmon) instead hinders the movement of F155 (red) by blocking the necessary motions of neighbouring amino acids, which now act as physical barriers, preventing rotation. By blocking the rotation of F155, ORG275769 makes it more difficult for CB1R to change shape, reducing the ability of agonists to promote signalling.
In summary
This elegant series of structures and simulations convincingly illustrates the likely mechanism for allosteric modulation of CB1R by these compounds. The cleft where these modulators bind acts like a balance, with F155 at the centre, allowing these compounds to enhance or diminish signalling by literally pushing the receptor towards, or away from, shapes that enable activation. With this knowledge, drug discovery scientists can develop new modulators, suitable for use in humans, to alter CB1R activity more gently for therapeutic effect.
